Contents
semPlot
semPaths
# A silly dataset:
X <- rnorm(100)
Y <- rnorm(100)
Z <- rnorm(1) * X + rnorm(1) * Y + rnorm(1) * X * Y
DF <- data.frame(X, Y, Z)
# Regression including interaction:
res <- lm(Z ~ X * Y, data = DF)
# Path diagram:
semPaths(res)
# Standardized estimates:
semPaths(res, "std", "hide")
MIMIC model
Lavaan
library("lavaan")
# Example 5.8 from mplus user guide:
Data <- read.table("http://www.statmodel.com/usersguide/chap5/ex5.8.dat")
names(Data) <- c(paste("y", 1:6, sep = ""), paste("x", 1:3, sep = ""))
# Model:
model.Lavaan <- "f1 =~ y1 + y2 + y3\nf2 =~ y4 + y5 + y6\nf1 + f2 ~ x1 + x2 + x3 "
# Run Lavaan:
library("lavaan")
fit <- lavaan:::cfa(model.Lavaan, data = Data, std.lv = TRUE)
# Plot path diagram:
semPaths(fit, title = FALSE, curvePivot = TRUE)
# Standardized parameters:
semPaths(fit, "std", edge.label.cex = 0.5, curvePivot = TRUE)
Mplus
# Same model, now using mplus output:
download.file("http://www.statmodel.com/usersguide/chap5/ex5.8.out", outfile <- tempfile(fileext = ".out"))
# Plot model:
semPaths(outfile, intercepts = FALSE)
## Reading model: /tmp/RtmpOqhQqG/file3e0f6696510b.out
# Note that mplus did not report the fixed variances of the exogenous
# variables.
Thresholds
Lavaan
# Example 5.8 from mplus user guide:
Data <- read.table("http://www.statmodel.com/usersguide/chap5/ex5.2.dat")
names(Data) <- c("u1", "u2", "u3", "u4", "u5", "u6")
Data <- as.data.frame(lapply(Data, ordered))
# Lavaan model:
model <- " f1 =~ u1 + u2 + u3; f2 =~ u4 + u5 + u6 "
# Run Lavaan:
fit <- lavaan::cfa(model, data = Data)
# Plot path diagram:
semPaths(fit, intercepts = FALSE)
Mplus
# Same model, now using mplus output:
download.file("http://www.statmodel.com/usersguide/chap5/ex5.2.out", outfile <- tempfile(fileext = ".out"))
# Plot model:
semPaths(outfile)
## Reading model: /tmp/RtmpOqhQqG/file3e0f3be9b0b8.out
OpenMx
# To isntall OpenMx see: http://openmx.psyc.virginia.edu/
library("OpenMx")
# Example from mxRun help page: Create and run the 1-factor CFA on the
# openmx.psyc.virginia.edu front page
data(demoOneFactor) # load the demoOneFactor dataframe
manifests <- names(demoOneFactor) # set the manifest to the 5 demo variables
latents <- c("G") # define 1 latent variable
model <- mxModel("One Factor", type = "RAM", manifestVars = manifests, latentVars = latents,
mxPath(from = latents, to = manifests), mxPath(from = manifests, arrows = 2),
mxPath(from = latents, arrows = 2, free = FALSE, values = 1), mxData(cov(demoOneFactor),
type = "cov", numObs = 500))
model <- mxRun(model) #run model, returning the result
## Running One Factor
# Plot with colors from OpenMx front page:
semPaths(model, color = list(lat = rgb(245, 253, 118, maxColorValue = 255),
man = rgb(155, 253, 175, maxColorValue = 255)), mar = c(10, 5, 10, 5))
## Factor Analysis:
source("http://openmx.psyc.virginia.edu/svn/trunk/demo/TwoFactorModel_PathCov.R")
## Running Two Factor Model Path
## twoFactorFit@output$estimate[["l2"]] and 0.972 are equal to within 0.01.
## twoFactorFit@output$estimate[["l3"]] and 0.931 are equal to within 0.01.
## twoFactorFit@output$estimate[["l5"]] and 1.0498 are equal to within 0.01.
## twoFactorFit@output$estimate[["l6"]] and 1.0533 are equal to within 0.01.
## twoFactorFit@output$estimate[["varF1"]] and 0.6622 are equal to within 0.01.
## twoFactorFit@output$estimate[["varF2"]] and 0.451 are equal to within 0.01.
## twoFactorFit@output$estimate[["cov"]] and 0.2958 are equal to within 0.01.
## twoFactorFit@output$estimate[["e1"]] and 0.3348 are equal to within 0.01.
## twoFactorFit@output$estimate[["e2"]] and 0.3994 are equal to within 0.01.
## twoFactorFit@output$estimate[["e3"]] and 0.4101 are equal to within 0.01.
## twoFactorFit@output$estimate[["e4"]] and 0.542 are equal to within 0.01.
## twoFactorFit@output$estimate[["e5"]] and 0.4809 are equal to within 0.01.
## twoFactorFit@output$estimate[["e6"]] and 0.5586 are equal to within 0.01.
## twoFactorFit@output$estimate[["meanx1"]] and 2.988 are equal to within 0.01.
## twoFactorFit@output$estimate[["meanx2"]] and 3.011 are equal to within 0.01.
## twoFactorFit@output$estimate[["meanx3"]] and 2.986 are equal to within 0.01.
## twoFactorFit@output$estimate[["meany1"]] and 2.955 are equal to within 0.01.
## twoFactorFit@output$estimate[["meany2"]] and 2.956 are equal to within 0.01.
## twoFactorFit@output$estimate[["meany3"]] and 2.967 are equal to within 0.01.
semPaths(twoFactorFit, layout = "tree2")
Multi-group analysis
## LISREL: Download measurment invariance example:
download.file("http://sachaepskamp.com/files/mi1.OUT", modFile <- tempfile(fileext = ".OUT"))
layout(t(1:2))
semPaths(modFile, "eq", ask = FALSE, as.expression = "edges", mar = c(3, 1,
5, 1))
# Color indicates equality constraints.
semSyntax
# MIMIC model, example 5.8 from mplus user guide:
Data <- read.table("http://www.statmodel.com/usersguide/chap5/ex5.8.dat")
names(Data) <- c(paste("y", 1:6, sep = ""), paste("x", 1:3, sep = ""))
# Data <- Data[,c(7:9,1:6)]
# Model:
model.Lavaan <- "f1 =~ y1 + y2 + y3\nf2 =~ y4 + y5 + y6\nf1 + f2 ~ x1 + x2 + x3 "
# Run Lavaan:
library("lavaan")
fit.Lavaan <- lavaan:::cfa(model.Lavaan, data = Data, std.lv = TRUE)
# Obtain Lavaan syntax:
model.Lavaan2 <- semSyntax(fit.Lavaan, "lavaan")
##
## Model <- '
## f1 =~ y1
## f1 =~ y2
## f1 =~ y3
## f2 =~ y4
## f2 =~ y5
## f2 =~ y6
## f1 ~ x1
## f1 ~ x2
## f1 ~ x3
## f2 ~ x1
## f2 ~ x2
## f2 ~ x3
## y1 ~~ y1
## y2 ~~ y2
## y3 ~~ y3
## y4 ~~ y4
## y5 ~~ y5
## y6 ~~ y6
## f1 ~~ 1*f1
## f2 ~~ 1*f2
## f1 ~~ f2
## x1 ~~ 2.92060364355652*x1
## x1 ~~ 0.175914393445822*x2
## x1 ~~ -0.150978787074975*x3
## x2 ~~ 2.08178484352461*x2
## x2 ~~ 0.125476432507783*x3
## x3 ~~ 1.04155105145699*x3
## '
# Run Lavaan again:
fit.Lavaan2 <- lavaan:::lavaan(model.Lavaan2, data = Data)
# Compare models:
layout(t(1:2))
semPaths(fit.Lavaan, "std", title = FALSE)
title("Lavaan model 1", line = 3)
semPaths(fit.Lavaan2, "std", title = FALSE)
title("Lavaan model 2", line = 3)
# Convert to sem model:
model.sem <- semSyntax(fit.Lavaan, "sem")
##
## Model <- specifyModel()
## f1 -> y1,par1,NA
## f1 -> y2,par2,NA
## f1 -> y3,par3,NA
## f2 -> y4,par4,NA
## f2 -> y5,par5,NA
## f2 -> y6,par6,NA
## x1 -> f1,par7,NA
## x2 -> f1,par8,NA
## x3 -> f1,par9,NA
## x1 -> f2,par10,NA
## x2 -> f2,par11,NA
## x3 -> f2,par12,NA
## y1 <-> y1,par13,NA
## y2 <-> y2,par14,NA
## y3 <-> y3,par15,NA
## y4 <-> y4,par16,NA
## y5 <-> y5,par17,NA
## y6 <-> y6,par18,NA
## f1 <-> f1,NA,1
## f2 <-> f2,NA,1
## f1 <-> f2,par19,NA
## x1 <-> x1,NA,2.92060364355652
## x1 <-> x2,NA,0.175914393445822
## x1 <-> x3,NA,-0.150978787074975
## x2 <-> x2,NA,2.08178484352461
## x2 <-> x3,NA,0.125476432507783
## x3 <-> x3,NA,1.04155105145699
# Run sem:
library("sem")
fit.sem <- sem:::sem(model.sem, data = Data)
# Compare models:
layout(t(1:2))
semPaths(fit.Lavaan, "std", title = FALSE)
title("Lavaan", line = 3)
semPaths(fit.sem, "std", title = FALSE)
title("sem", line = 3)
lisrelModel
## Example of a Full LISREL model path diagram with the same number of
## exgenous and endogenous variables:
# Lambda matrices:
Loadings <- rbind(diag(1, 2, 2), diag(1, 2, 2), diag(1, 2, 2))
# Phi and Psi matrices:
LatVar <- diag(1, 2, 2)
# Beta matrix:
Beta <- matrix(0, 2, 2)
Beta[1, 2] <- 1
# Theta matrices:
ManVar <- diag(1, nrow(Loadings), nrow(Loadings))
# Gamma matrix:
Gamma <- diag(1, 2, 2)
# Tau matrices:
ManInts <- rep(1, 6)
# Alpha and Kappa matrices:
LatInts <- rep(1, 2)
# Combine model:
mod <- lisrelModel(LY = Loadings, PS = LatVar, BE = Beta, TE = ManVar, LX = Loadings,
PH = LatVar, GA = Gamma, TD = ManVar, TY = ManInts, TX = ManInts, AL = LatInts,
KA = LatInts)
# Plot path diagram:
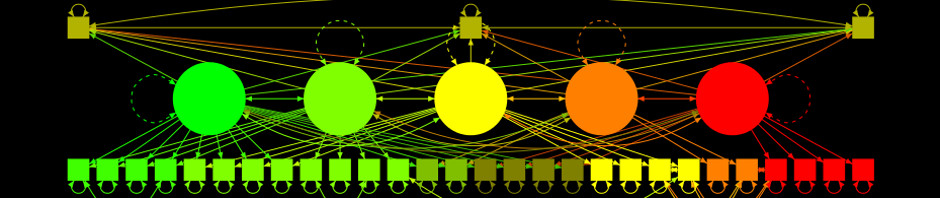
semPaths(mod, as.expression = c("nodes", "edges"), sizeMan = 3, sizeInt = 1,
sizeLat = 4)
# Plot path diagram with more graphical options:
semPaths(mod, as.expression = c("nodes", "edges"), sizeMan = 3, sizeInt = 1,
sizeLat = 4, label.prop = 0.5, curve = 0.5, bg = "black", groups = "latents",
intercepts = FALSE, borders = FALSE, label.norm = "O")
tricks
# A silly dataset:
A <- rnorm(100)
B <- A + rnorm(100)
C <- B + rnorm(100)
DF <- data.frame(A, B, C)
# Two regressions:
res1 <- lm(B ~ C, data = DF)
res2 <- lm(A ~ B + C, data = DF)
# Plot both in the same path diagram in two ways:
semPaths(res1 + res2, "model", "est", intercepts = FALSE)
semPaths(list(res1, res2), "model", "est", intercepts = FALSE)
semPlotModel-class
showClass("semPlotModel")
## Class "semPlotModel" [package "semPlot"]
##
## Slots:
##
## Name: RAM Vars Thresholds Computed ObsCovs ImpCovs
## Class: data.frame data.frame data.frame logical list list
##
## Name: Original
## Class: list